Research - (2024) Volume 19, Issue 2
Received: 03-Jun-2024, Manuscript No. gpmp-24-141068; Editor assigned: 04-Jun-2024, Pre QC No. P-141068; Reviewed: 17-Jun-2024, QC No. Q-141068; Revised: 21-Jun-2024, Manuscript No. R-141068; Published: 28-Jun-2024
Diabetes is a diverse and intricate metabolic illness defined by high levels of glucose in the blood due to either resistance to insulin, inadequate production of insulin, or both. Metformin is one of the initial oral hypoglycemic agents frequently prescribed. Toll-Like 4 Receptors (TLR4) are receptors located on the surface of cells. It triggers the body's natural immune responses to harmful microorganisms by initiating a series of biochemical reactions that activate kinases and transcription factors. Two significant non-synonymous Single Nucleotide Polymorphisms (SNPs) (rs4986790 and rs4986791) have been discovered in the coding region (exon 3) of the TLR4 gene.
Methodology: This prospective cohort study included one hundred patients who were newly diagnosed with type 2 diabetes mellitus. The study was carried out at Al Diwaniyah Teaching Hospital, Diwaniya Governorate/ Iraq. The age varied, with a mean age of 55.25 and a standard deviation of 9.88. There was a 12-week interval between the first blood sample (taken at diagnosis when therapy was not yet underway) and the second (taken 12 weeks later) for every patient.
Results: After administering metformin medication, there was a notable decrease in the average BMI (p<0.01). The levels of fasting plasma glucose, plasma insulin, HbA1c, and insulin resistance (measured by the HOMA-IR index) were considerably decreased (p< 0.001), while there was a significant increase in insulin sensitivity (measured by the QUICKI) (p< 0.001). There was a substantial decrease in the average levels of lipid profiles (p< 0.001), except for an increase in the average HDL level.
There was no significant linkage seen between TLR4 gene variant Asp299Gly and glycemic control, as measured by HbA1c levels below 7%, after therapy with metformin (p=0.256).
Conclusion: Metformin has demonstrated efficacy in enhancing serum lipid profiles, insulin levels, fasting plasma glucose, HbA1c, the insulin resistant index (HOMA-IR), and the insulin sensitivity index (QUICKI) in individuals diagnosed with type 2 diabetes mellitus. However, the TLR4 gene Asp299Gly polymorphism does not significantly correlate with glycemic control or response to metformin treatment.
Polymorphism; TLR-4; Asp299Gly; Metformin
Diabetes mellitus refers to a variety of different medical conditions that all include metabolic abnormalities resulting in high blood sugar levels. These abnormalities might be caused by a decrease in insulin secretion, a resistance to insulin's effects, or both [1].
Type 2 Diabetes Mellitus (T2DM) is a frequently prevalent clinical manifestation and is commonly observed in adults aged 35 years and older. T2DM is marked by the presence of insulin resistance along with normal or excessive insulin secretion in the early stages. However, as time progresses, patients experience dysfunction of the beta cells and insufficient insulin production.
Consequently, some patients may need to be treated with insulin to regulate their blood glucose levels in the later stages of the disease [2]. T2DM is prevalent both worldwide and in Iraq [3]. Obesity is a prominent risk factor [4], and genetics play a substantial role in the development of the illness [5]. The management of diabetes mellitus necessitates the reduction of body weight, the implementation of nutritional interventions, the adoption of lifestyle adjustments, and the use of medication [6].
Metformin, a biguanide medicine, is a primary pharmacological agent used to manage the condition. It acts by inhibiting hepatic glucose synthesis, lowering intestinal glucose absorption, and increasing insulin sensitivity in target tissues, leading to decreased blood glucose levels. Metformin reduces both the fasting and after-meal blood glucose levels. Metformin is frequently employed as a standalone treatment or in conjunction with other therapies when dietary adjustments and physical activity fail to sufficiently reverse hyperglycemia. According to the American Diabetes Association, metformin is the favored medication for newly diagnosed patients with T2DM who are adults or children aged ten and above [7-9]. Due to its efficacy, safety, affordability, and widespread usage, it is highly recommended as the initial treatment for T2DM [10]. Nevertheless, there is variation among individuals in their reaction to metformin as a therapeutic treatment. Research has demonstrated that commonly occurring genetic variations have an impact on the way metformin affects blood sugar levels, accounting for up to 34% of the differences observed in the decrease of hemoglobin A1c (HbA1c) levels while using this medication [11]. The pharmacokinetic gene variations in candidate gene studies have not consistently and significantly influenced the glycemic response to metformin in individuals with T2DM [12]. In addition, only a limited number of genome-wide signals linked to the clinical response to metformin have been identified thus far [11,13].
Numerous diseases, such as diabetes, cancer, arthritis, and cardiovascular issues, have long been linked to inflammation. Important components of the innate immune system, Toll-Like Receptors (TLRs) identify conserved pathogen motifs, also known as Pathogen-Associated Molecular Patterns (PAMPs), as well as Damage-Associated Molecular Patterns (DAMPs), which are endogenous host materials that are propagated during cellular stress or death.
In the coding region (exon 3) of the TLR4 gene, two significant non-synonymous single nucleotide polymorphisms, rs4986790 (Asp299Gly) (A/G) and rs4986791 (Thr399Ile) (C/T), have been found, which are in a tight linkage disequilibrium [14]. Although some research has linked TLR4 polymorphisms to an increased risk of diabetes, Research has also shown that bearers of the Asp299Gly and Thr399Ile genotypes significantly reduce the incidence of diabetes complications. There was no significant association between TLR4 polymorphism and diabetes or diabetes complications, according to other research [15].
This prospective cohort study included one hundred patients who were recently diagnosed with T2DM (52 males and 48 females). The study was worked on starting in December 2022 and ending in August 2023. The study was done in the Al-Diwaniyah teaching hospital. An endocrinologist made the diagnosis for the enrolled T2DM patients.
The age varied, with a mean age of 55.25 and a standard deviation of 9.88. There was a 12-week interval between the first blood sample (taken at diagnosis when therapy was not yet underway) and the second (taken 12 weeks later) for every patient.
Phenotypic analysis: Data on age, gender, body mass index, smoking history, familial association with T2DM, and chronic medications were collected for each patient. A blood sample was collected and divided into two parts. One part was used to test fasting blood sugar, serum lipid profile, and serum insulin levels, while the other part was utilized for genetic analysis of TLR-4 gene. The HbA1c levels were determined using the Finecare™ HbA1c (Hemoglobin A1c) Rapid Quantitative Test. The serum measures were taken at two time points: baseline and 3 months following the start of metformin.
Genotyping analysis: The DNA was extracted from the blood samples of all T2DM patients by using the Favor Prep™ Blood Genomic DNA kit (Favorgen). The DNA concentration and purity of the samples were assessed using a nanodrop device.
A Polymerase Chain Reaction (PCR) was performed using a specific section of the genome, amplified using a thermocycler program called T-professional, manufactured by Biometra (Germany). The primers were provided by Alpha-DNA in the form of a lyophilized powder. The primer sets utilized in this investigation are displayed in Tab. 1. The 2x Taq plus PCR smart mix kit from SolGent (South Korea) was employed.
| SNPs | Primers | Amplicon size (bp) |
|---|---|---|
| Asp299Gly rs4986790 |
F: 5’ -GAT TAG CAT ACT TAG ACT ACT ACC TCC ATG-3’ R: 5’-GAT CAA CTT CTG AAAA GCA TTC CCAC-3’ |
A: 249 G: 223 |
Tab. 1. Sequence of primers for detecting the TLR4 SNP.
The condition was fine-tuned to optimize the reaction necessary for DNA amplification. The PCR technique utilized in this investigation commenced with an initial cycle at a temperature of 95 °C for a length of 5 minutes, followed by 30 consecutive cycles. The cycle involved the primers being denatured at 95 °C for 30 seconds, followed by annealing at 60 °C for 30 seconds, and extension at 72 °C for 30 seconds. The reaction was concluded by exposing it to a final synthesis step at a temperature of 72 °C for duration of 10 minutes. The amplicon resulting from the amplification reaction was cut at a specified site using restriction endonuclease using the RFLP technique. Specifically, Nco I restriction enzyme (10 U/l) was used for Asp299Gly, which was incubated at 37 °C for 1 hour. This enzyme was obtained from Promega Corporation. This cutting process allows for further analysis of the amplicon. The digested product was separated using 2% agarose gel electrophoresis (Condalab, Canada), which was stained with diamond nucleic acid dye that was used for coloring the gel for observation by UV apparatus.
Statistical analysis: SPSS 26 and Excel 2019 were used to gather, summarize, analyze, and present data. The means were compared before and after therapy using a paired t-test. When comparing means across many groups, the Kruskal-Wallis test was employed. All of the genes that were analyzed were evaluated for Hardy-Weinberg equilibrium, and a chi-square test was used to examine the use of categorical variables. Statistical significance was defined as<0.05.
The present study comprised a cohort of 100 individuals diagnosed with T2DM who had not yet been treated with any pharmaceutical interventions. The patient population consisted of 52 males (52%), and 48 females (48%). A total of 59 patients reported a familial history of diabetes mellitus. After receiving metformin medication, there was a notable decrease in the average BMI (p<0.01). A significant reduction in fasting plasma glucose, HbA1c, insulin level, and insulin resistance was observed before and after therapy with metformin. Furthermore, there was a substantial increase in insulin sensitivity as measured by the QUICKI (p<0.001). There was a substantial decrease in the average levels of serum triglyceride, total cholesterol, VLDL, and LDL before and after therapy with metformin, while there was a notable rise in serum HDL levels both before and after therapy with metformin (p< 0.0001), as shown in Tab. 2.
| Descriptive Statistics | Baseline Mean ± SD n=100 |
After treatment Mean ± SD n=100 |
P |
|---|---|---|---|
| BMI (kg/m2) | 33.51 ± 8.74 | 30.66 ± 7.70 | <0.01 |
| FPG (mg/dl) | 245.52 ± 87.57 | 156.91 ± 70.02 | <0.0001 |
| HbAIc | 9.315 ± 1.60 | 7.00 ± 1.06 | <0.0001 |
| Plasma insulin mU/L | 22.782 ± 7.43 | 15.78 ± 7.6 | <0.0001 |
| HOMA-IR index | 13.81 ± 6.49 | 6.17 ± 4.50 | <0.0001 |
| QUICKI | 0.271 ± 0.014 | 0.3 ± 0.03 | <0.0001 |
| Triglyceride (mg/dl) | 255 ± 71.2 | 187 ± 70.2 | <0.0001 |
| Cholesterol (mg/dl) | 219 ± 41 | 179 ± 30 | <0.0001 |
| HDL (mg/dl) | 37 ± 7.5 | 42.87 ± 7.6 | <0.0001 |
| VLDL (mg/dl) | 51.1 ± 12.94 | 41.3 ± 14.2 | <0.0001 |
| LDL (mg/dl) | 130 ± 41 | 92.16 ± 40.3 | <0.0001 |
Tab. 2. Comparison of glycemic parameters and serum lipid profile before and after treatment with metformin.
The allele frequency of the SNP in the study is shown in Tab. 3. After PCR amplification, the product of the TLR-4 Asp299Gly A/G was digested via NcoI restriction enzyme, 2% agarose gel was used to analyze the product which yielded bands according to Tab. 1. The results are presented in Fig. 1.
| SNPs | Percentage | Allele Frequency |
|---|---|---|
| Asp299Gly (rs4986790) |
||
| AA | 65 | A=0.8 |
| AG | 30 | G=0.2 |
| GG | 5 | - |
Tab. 3. Genotype distribution of study groups.
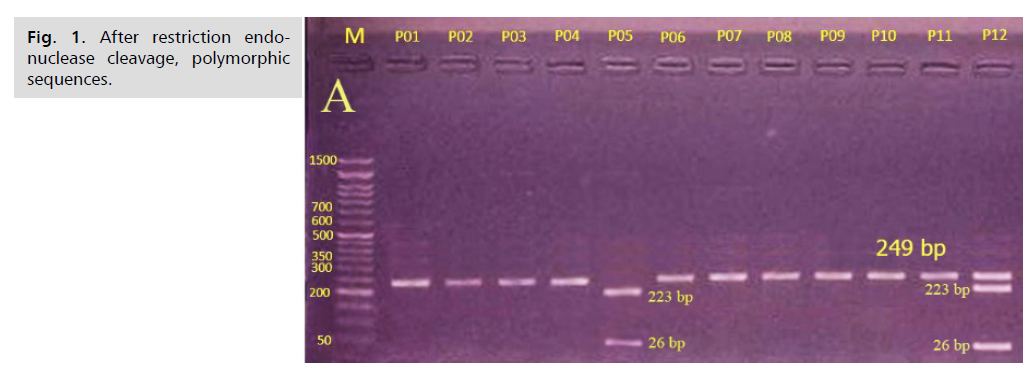
Fig 1. After restriction endonuclease cleavage, polymorphic sequences.
The observed and expected frequencies of T2DM patients for Asp299Gly were not significantly different from Hardy-Weinberg equilibrium (p=0.8226). To assess the impact of the Asp299Gly variation in gene on patients glycemic response to treatment with metformin, we compared mean HbA1c levels after treatment according to the gene polymorphism, and the results showed no significant difference in mean HbA1c (p=0.256).
A comparison of phenotypic characteristics, including glycemic parameters and lipid profiles, before and after metformin treatment according to Asp299Gly gene polymorphisms in the co-dominant and dominant models showed there were no significant differences relevant to the variant allele in any of these phenotypic characteristics (p>0.05).
Based on the estimates of the World Health Organization, by 2030, diabetes mellitus is expected to rank as the seventh most common cause of death globally [16]. Overall, metformin is very effective and stands out among other oral treatments for T2DM due to its ability to encourage weight reduction instead of weight gain. Although metformin has several advantageous characteristics, it is not a universal remedy. Empirical evidence from clinical practice and corroborating studies indicates that metformin monotherapy often falls short in attaining glycemic control for around 21% of individuals who are on this medication. Therapeutic interventions do not achieve desired levels of plasma glucose control during the first 5 years of treatment [17].
The heterogeneity in response to metformin is presumably influenced by hereditary factors, which contribute to individual variability. After administering metformin as part of this trial, there was a notable decrease in the average BMI (p<0.01). The Diabetes Prevention Program (DPP) is the most extensive research project demonstrating the weight advantages of metformin. Of the individuals included in the present investigation, 59.0% revealed a family history of diabetes mellitus. Individuals who have a family history of T2DM in a first-degree relative are 2 to 3 times more likely to acquire T2DM compared to those without such a family history [18,19]. Scott, et al., found that those with a positive family history of diabetes mellitus had a 2.7 times higher chance of developing T2DM compared to those without a family history of diabetes [20]. In the current study, there was also a significant reduction in fasting plasma glucose, HbA1c, plasma insulin, and insulin resistance, as represented by the HOMO-IR index. The findings are consistent with earlier studies and are directly connected to the mechanism of action of metformin [21,22].
The present research found that although HDL-C levels increased, mean blood triglyceride, total cholesterol, VLDL-C, and LDL levels significantly decreased in newly diagnosed T2DM patients after 3 months of using metformin. Metformin has been proven to significantly lower triglyceride and LDL-C and raise HDL-C in a cohort trial that included 155 newly diagnosed T2DM [23]. Metformin was demonstrated in another cross-sectional study to significantly lower total cholesterol and LDL-C levels in 150 newly diagnosed diabetes patients treated for three months [24]. In the current study, analysis for BMI, glycemic parameters, and serum lipid profile values was conducted by the Kruskal-Wallis test in relation to the genotype of Asp299Gly of the TLR-4 gene.
The co-dominant model revealed no significant relationship for phenotypic parameter analysis; means did not show significant modifications before and following treatment with metformin in newly diagnosed T2DM patients, as well as under the dominant pattern. Many prior clinically significant studies in pharmacogenomics have included genes that have shown an association between TLR-4 Asp299Gly SNP and a reduced risk of developing atherosclerosis and T2DM [25]. Several investigations have shown a correlation between TLR-4 polymorphism and T2DM [26]. There was no relationship between TLR-4 polymorphism and diabetes or its consequences, according to another research. [27]. In their meta-analysis, Yin, et al. found no association between the TLR4 gene Asp299Gly and Thr399Ile polymorphisms and an elevated risk of T2DM [28]. In many inflammatory contexts, recent research has shown that TLR4 expression may be correlated with AMPK signaling. Multiple models of inflammatory diseases have shown that AMPK, which acts as a detector of cellular energy, has anti-inflammatory properties [29]. A recent animal study has shown that metformin minimizes the activity of the TLR4 inflammatory pathway in the muscular tissues of diabetic rats. The findings suggest a connection between TLR4 and AMPK levels, which indicates the potential of metformin to attenuate TLR4 pathway's activation. These modifications may explain the increase in insulin sensitivity reported in diabetic rats administered metformin [30].
The findings indicated that metformin had significantly favorable impacts on fasting plasma glucose, HbA1c level, serum insulin level, insulin resistance index (HOMA-IR), and insulin sensitivity index (QUICKI) in individuals recently diagnosed with T2DM. Similarly, metformin had a significantly positive impact on lipid profiles, with a notable decrease in all levels except for HDL, which showed a considerable rise. Additionally, metformin had an impact on BMI, resulting in a considerable decrease. The TLR4 gene polymorphism Asp299Gly did not have a significant influence on the therapeutic response of metformin, according to the analysis using the co-dominant and dominant models.
Approval from the Ethical Committee (in the faculty of pharmacy at Kufa University) was obtained for the protocol of the study.
There are no conflicts of interests.
The source of this research comes from myself.
(A) Study Design · (B) Data Collection · (C) Statistical Analysis · (D) Data Interpretation · (E) Manuscript Preparation · (F) Literature Search · (G) No Fund Collection
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Copyright:This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.