Research Article - (2024) Volume 19, Issue 2
Molecular characterization of third generation cephalosporin resistant in E. coli isolated from urinary tract infection
Khamael Arif Mahdi Al-challabi1, Wurood Kadhim Abed2 and Ali Razzaq Hussein3*Received: 01-Jun-2024, Manuscript No. gpmp-24-142311; Editor assigned: 03-Jun-2024, Pre QC No. P-142311; Reviewed: 15-Jun-2024, QC No. Q-142311; Revised: 21-Jun-2024, Manuscript No. R-142311; Published: 28-Jun-2024
Abstract
Background:Urinary Tract Infections (UTIs) are quite possibly of the most widely recognized bacterial problems around the world, affecting many individuals in both sex, especially women, so UTI is related to Urogynecology. The most common bacteria that causes UTIs is uropathogenic E. coli. Although Third Generation Cephalosporins (3GCs) have proven to be efficacious in treating UTIs, there is a rise in antibiotic resistance presents a serious risk to public health.
Aim: The reason of this work is to determine which genes make E. coli resistant to 3GCs.
Methods: One hundred urine samples were gathered from hospitalized patients and outpatients who had Urinary Tract Infections (UTIs) visiting Balad general hospital, Saladin governorate, from November 2023 until March 2024, the specimens were cultured on MacConkey and positive pure growths of E. coli were confirmed by API 20 after that subjected to disc diffusion testing against six different antibiotics then screened against antibiotic resist ESBLS (blaTEM, blaSHV, blaOXA and blaCTX-M) genes.
Results: most of UTI cases were female (85/100, 85%). Twenty-four (24) E. coli isolates were obtained from UTIs, the majority of the E. coli isolates were found to be resistant to antibiotics, as indicated by the antibiogram results: 100% to Cefoperazone, 95.8% to cefotaxime, 86.7 to ceftriaxone, 83.3% to cefexime, 83.3% to cefpodoxime, and 83.2% to ceftazidime. The consequences of the study uncovered those 21 (87.5%) isolates contained blaTEM gene, same for blaSHV, 22 (91.6%) isolates contained blaOXA gene, and 20 (83.3%) isolates contained blaCTX-M gene.
Conclusion: The current study confirms the prevalence of multidrug resistance E. coli and coexistence of ESBLS genes in most E. coli isolated from UTI patient, which most of them are females.
Keywords
Cephalosporin; 3GCs; Antibiotic resistance; UTI; E. coli
Introduction
UTIs, or urinary tract infections, are among the most prevalent bacterial genecology/urogynaecology diseases in the world, impacting millions of people each year, mostly women. The most common infection that causes illness is E. coli, responsible for up to 80-90% of uncomplicated UTIs. The management of UTIs has become increasingly challenging due to the rising prevalence of antimicrobial resistance, particularly among uropathogenic E. coli strains [1]. Third-Generation Cephalosporins (3GCs), such as ceftriaxone and ceftazidime, have historically been effective in treating UTIs, but the emergence of resistance to these antibiotics represents a huge public health threat [2].
Resistance to 3GCs in E. coli isolates is often mediated by various molecular mechanisms that confer resistance to β-lactam antibiotics. The synthesis of Extended-Spectrum Β-Lactamases (ESBLs), which are enzymes that can hydrolyse a variety of β-lactam antibiotics, for example, monobactams, cephalosporins, and penicillins, is one of the main processes [3]. Multiple antibiotic classes are commonly resistant to E. coli strains that produce ESBL, limiting treatment options and increasing the risk of treatment failure and recurrent infections [4]. In addition to ESBLs, another important mechanism of 3GC resistance in E. coli is the production of AmpC β-lactamases, which are chromosomally encoded enzymes capable of hydrolyzing cephalosporins and monobactams [5]. AmpC-producing E. coli strains often exhibit resistance to multiple β-lactam antibiotics, such as combinations of β-lactam/β-lactamase inhibitors and third-generation cephalosporins. Furthermore, alterations in outer membrane porins, such as OmpF and OmpC, can contribute to decreased susceptibility to β-lactam antibiotics by reducing the influx of these drugs into bacterial cells [6]. Additionally, overexpression of efflux pump systems, which actively extrude antibiotics from bacterial cells, can confer resistance to multiple antibiotic classes, including β-lactams [7].
Understanding the molecular characteristics of 3GC-resistant E. coli isolates from UTIs is essential for guiding empirical antibiotic therapy, implementing infection control measures, and developing strategies to combat antimicrobial resistance. The purpose of this work is to look into the mechanisms and genetic determinants of 3GC resistance in E. coli isolates from UTIs through molecular characterization techniques, such as Polymerase Chain Reaction (PCR). By elucidating the molecular basis of antibiotic resistance in UTI-causing E. coli strains, this research endeavors to provide valuable insights into the epidemiology, pathogenesis, and management of antibiotic-resistant UTIs, ultimately contributing to the improvement of more successful remedial interventions and antimicrobial stewardship initiatives. The goal of the present study was to identify E. coli that was resistant to medicines among UTI patients who visited Balad General Hospital.
Methodology
Sample collection and identification: A total of 100 urine specimens taken from patients with urinary tract infections both outpatient and inpatient (UTIs) visiting Balad general hospital, Saladin governorate, from November 2023 until March 2024. Five to ten millilitres of midstream pee were provided by each patient, and the sample was collected in clean, sterile, leak-proof bottles. The subjects were then given the appropriate instructions, and the MSU was then collected into a sterile, wide-mouthed urine container. The gathered samples were placed on MacConkey agar, inoculated, and left to develop bacteria throughout the entire night at 37 °C. The urine sample with pure E. coli isolates were chosen for additional testing by API (Analytical Profile Index) 20E strip to confirm identification of E. coli. For additional molecular analysis, these isolates were kept at -80 °C in Tryptone Soya Broth (TSB) media containing 15% glycerol [8].
Antimicrobial susceptibility testing: Using E. coli ATCC 25922 as quality control strains, antimicrobial susceptibility testing was conducted using the disk diffusion technique in accordance with the Kirby-Bauer method. To test for 3GC-resistant E. coli isolates, a calibrated bacterial suspension (0.5 McFarland) was inoculated onto MHA that had been enhanced with antibiotics. In light of this, from an agar plate culture, three to five well-isolated colonies of the similar morphological sort were picked, put into Muller Hinton broth, and refined for 24 hours at 37 °C. To get the suspension's turbidity visually equivalent to the 0.5 McFarland standards, the turbidity was corrected using sterile saline. The freshly produced Mueller Hinton agar plate was then completely covered with the swab. Within fifteen minutes, the antimicrobial disks were placed on the plates after inoculation. After that, the plates were incubated for 24 hours at 37°C. Based on the interpretation of resistance data, a zone of inhibition was assessed, and the results were classified as sensitive, resistant, or intermediate by the Clinical and Laboratory Standards Institute (CLSI, 2021). The antimicrobial agents tested were ceftriaxone (CRO, 30 μg), ceftazidime (CAZ, 30 μg), cefotaxime (CTX, 30 µg), Cefoperazone (CPZ, 75 µg), cefixime (CFX, 10 μg), and cefpodoxime (CPD, 10 μg).
DNA extraction: Thermoscientific Gene JET Genomic DNA Kit was used to extract DNA from old broth cultures that had been incubated for 24 to 48 hours. Following extraction, the DNA was examined by electrophoresis on 1% Gel. At -20 °C, the DNA was retained.
Molecular characterization of antibiotic-resistant genes: Using a traditional Polymerase Chain Reaction (PCR) machine (Labnet International, Inc. USA), the selected antibiotic-resistant genes were amplified. For 3GC-resistant E. coli isolates, the most regular 3GC resistance genes typically recognized and encoding the blaCTX-M, blaTEM, blaSHV, and blaOXA genes [9]. The particular primers were utilized for the amplifications of the mentioned genes are summarized in Tab. 1., the amplification condition in Tab. 2.
| Target | Primer Name | Primer Sequence (5´-3´) | Molecular Weight | References |
|---|---|---|---|---|
| blaTEM | TEM -F | ATGAGTATTCAACATTTCCG | 867 | Brinas L, et al. (2002) [29] |
| TEM- R | CTGACAGTTACCAATGCTTA | |||
| blaSHV | SHV-F | GGGTTATTCTTATTTGTCGC | 930 | Brinas L, et al. (2002) [29] |
| SHV-R | TTAGCGTTGCCAGTGGTC | |||
| blaOXA | OXA-F | GGCACCAGATTCAACTTTCAAG | 564 | Brinas L, et al. (2002) [29] |
| OXA-R | GACCCCAAGTTTCCTGTAAGTG | |||
| blaCTX-M | CTX-M-F | TCAAGCCTGCCGATGGT | 561 | Pitout JD, et al. (2004) [28] |
| CTX-M-R | TGATTCTCGCCGATCTG |
Tab. 1. Primers utilized in the study.
| Genes | Initial | Cycling Condition | Final Extension | Cycles | ||
|---|---|---|---|---|---|---|
| Denaturation | Denaturation | Annealing | Extension | |||
| blaTEM | 94 °C/1 min | 94 °C/1 min | 58 °C/1 min | 72 °C/1 min | 72 °C/10 min | 30 |
| blaSHV | 94 °C/1min | 94 °C/1min | 56 °C/1 min | 72 °C/1 min | 72 °C/10 min | 30 |
| blaOXA | 94 °C/10 min | 94 °C/40 sec | 60 °C/40 sec | 72 °C/1 min | 72 °C/5 min | 30 |
| blaCTX-M | 94 °C/4min | 94 °C/30sec | 63 °C/1min | 72 °C/1min | 72 °C/5min | 35 |
Tab. 2. PCR blaTEM, blaSHV, blaOXA, and blaCTX-M genes thermocycling conditions.
Gel electrophoresis: Bands were seen using a gel documentation system (BIO-RAD Gel DocTM XR+), and the amplicon was run on 2% agarose gel electrophoresis (Bioron, Life Sciences). By comparing the amplicon sizes with a 100 bp DNA ladder, the sizes were found.
Results
Age and gender of cases: The patient’s age was ranged between 20-70 years, moreover, the clinical cases of UTI 85/100 (85%) were female and 15/100 (15%) were male.
Antibiotic susceptibility test: A total of twenty-four E. coli isolates were obtained from UTIs, and these were chosen for the current investigation based on phenotypic techniques to determine their resistance profiles. Using the CLSI-2021 criteria, all E. coli isolates were tested using the disc diffusion method against six selected antibiotics (Fig. 1.). Most of the E. coli isolates were found to be resistant to antibiotics, as per the antibiogram results; 100% to Cefoperazone, 95.8% to cefotaxime, 86.7 to ceftriaxone, 83.3% to cefexime, 83.3% to cefpodoxime, and 83.2% to ceftazidime (Tab. 3.).
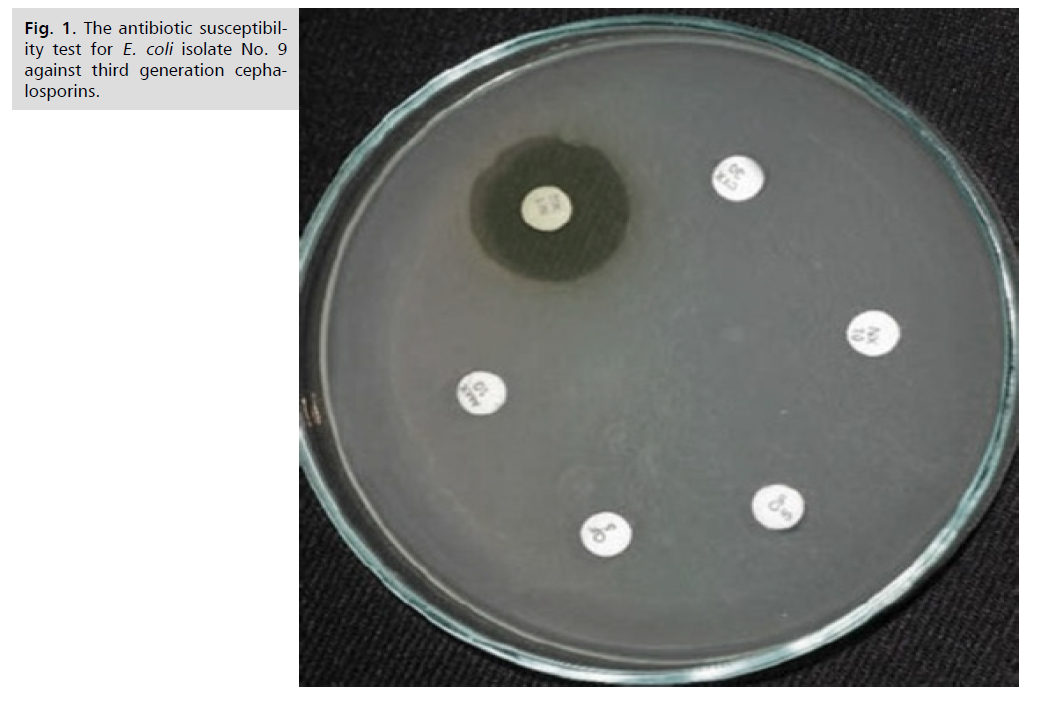
Fig 1. The antibiotic susceptibility test for E. coli isolate No. 9 against third generation cephalosporins.
| Antibiotic | Susceptibility Status | E. coli Isolates |
|---|---|---|
| Cefexime | R | 20 (83.3%) |
| S | 3 (12.5%) | |
| I | 1 (4.2%) | |
| Ceftriaxone | R | 21 (86.7%) |
| S | 3 (12.5%) | |
| I | - | |
| Cefotaxime | R | 23 (95.8%) |
| S | - | |
| I | 1 (4.2%) | |
| Cefoperazone | R | 24 (100.0%) |
| S | - | |
| I | - | |
| Cefpodoxime | R | 20 (83.3%) |
| S | 4 (16.7%) | |
| I | - | |
| Ceftazidime | R | 20 (83.2%) |
| S | 2 (8.4%) | |
| I | 2 (8.4%) |
Tab. 3. Antibiotic susceptibility profiles of E.coli isolates.
ESBLs gene screening: The isolates were tested for the existence of blaTEM, blaSHV, blaOXA, and blaCTX-M, four typical ESBL gene types. As shown in Fig. 2. the amplified PCR-products for the four genes in each of the two E. coli showed distinct molecular weights (867, 930, 564 and 561) bp, respectively. In the current investigation, clinical isolates of E. coli from patients with UTIs were used to characterize many targeted antibiotic resistance genes molecularly. The aftereffects of study uncovered those 21 (87.5%) isolates contained blaTEM gene, same for blaSHV, 22 (91.6%) isolates contained blaOXA gene, and 20 (83.3%) isolates contained blaCTX-M gene as shown in Tab. 4. Moreover, current study detected coexistence of studied ESBLS gene in two thirds of isolates.
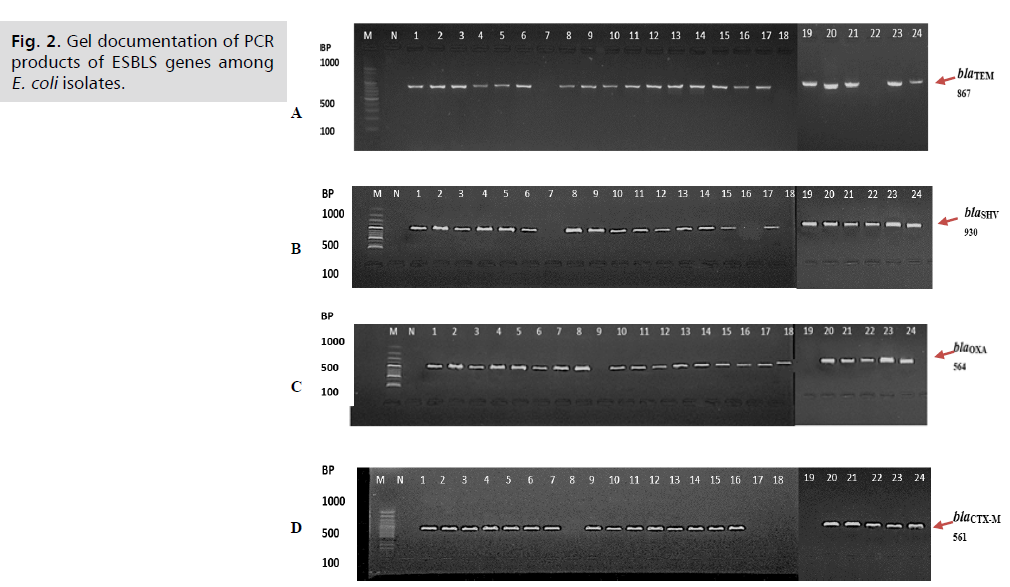
Fig 2. Gel documentation of PCR products of ESBLS genes among E. coli isolates.
| ESBL Gene Type | ESBL Gene Distribution | |
|---|---|---|
| Number | Percentage | |
| blaTEM | 21 | 87.50% |
| blaSHV | 21 | 87.50% |
| blaOXA | 22 | 91.60% |
| blaCTX-M | 20 | 83.30% |
| blaTEM + blaSHV | 20 | 83.30% |
| blaTEM + blaOXA | 19 | 79.10% |
| blaTEM + blaCTX-M | 18 | 75% |
| blaSHV + blaOXA | 19 | 79.10% |
| blaSHV + blaCTX-M | 18 | 75% |
| blaOXA + blaCTX-M | 19 | 79.10% |
| blaTEM + blaSHV + blaOXA | 19 | 79.10% |
| blaTEM + blaSHV + blaCTX-M | 18 | 75% |
| blaTEM + blaOXA + blaCTX-M | 18 | 75% |
| blaSHV + blaOXA + blaCTX-M | 17 | 70.80% |
| blaTEM + blaSHV + blaOXA + blaCTX-M | 16 | 66.60% |
Tab. 4. Distribution of ESBLs genes among E.coli isolates.
Discussion
The current study revealed higher UTI in female, there are many studies recorded that significantly lower prevalence of the bacteria implicated in UTI was observed in males as compared with females [10,11]. Every human gastrointestinal tract contains E. coli as part of the normal flora. However, on rare occasions, these bacteria can turn pathogenic and cause illnesses like bacteremia, wound infections, and UTIs [12]. UTIs can have negative consequences and are commonly caused by infections in both adults and children [13]. According to earlier research, E. coli is the primary bacterium responsible for UTIs.
It is known that members of the Enterobacteriaceae family, such as E. coli, can acquire both transferable and intrinsic resistance to antibiotics (via chromosomes and plasmids). This compels us to keep an eye out for AMR gene presence in bacterial isolates. Intrinsic resistance was attributed to broad-spectrum beta-lactamases such as blaTEM, blaSHV-1, and blaOXA-1 and blaCTX-M [14]. According to a study by Dirar MH, et al. [15], the most widely recognized genotypes in Enterobacteriaceae that produce ESBLs are blaTEM and blaCTX-M. Zaniani FR, et al. [16] determined that the frequency of blaSHV among the ESBLs generating E. coli was 14.4%. According to a study by Kiratisin P, et al. and colleagues [17], 11.8% of blaOXA was encoded by ESBL-producing E. coli.
According to Singh and his colleagues, blaTEM and blaCTX-M are highly predominant genes in E. coli isolates because conjugation assays of plasmid DNA revealed that they are plasmid-mediated and may spread between genera through Horizontal Gene Transfer (HGT). Jain P, et al. [18] conducted a study in Bangladesh and discovered that blaOXA (48%) and blaCTX-M (32%), two ESBL genes, were predominant in E. coli isolates that were used in the PCR analysis.
Antibiotics that prevent the production of cell walls fall within the beta-lactam class. A few examples of antibiotics with a beta-lactam ring are cephalosporins. However, by creating beta-lactamase enzymes, which catalyze beta-lactam antibiotics and so inactivate them, bacterial species have been able to counteract the actions of beta-lactam antibiotics. In light of this, researchers have created beta-lactamase inhibitors that, when paired with beta-lactam antibiotics, allow them to suppress the actions of bacterially produced beta-lactamases [19].
Bacteria have developed the capacity to withstand the effects of beta-lactam antibiotics over time. Antibiotics like cephalosporins were used to mediate infections caused by these bacteria. Additionally, certain bacterial species began exhibiting resistance to the class of medicines known as broad-spectrum cephalosporins. These bacteria were classified as species that produce ESBLs. It has been discovered that bacteria with genes like blaTEM become resistant to lesser cephalosporins like cefazolin and cephalothin. The presence of broad-spectrum cephalosporins with greater potencies, like cefotaxime, ceftriaxone, and ceftazidime, among others, hindered the growth of bacteria harboring blaTEM. Later, bacteria that carried the sulfhydryl variable (blaSHV) gene were discovered to have decreased resistance to broad-spectrum cephalosporins [20].
In the current investigation, blaCTX-M type ESBL was shown to be highly prevalent. Similar results were seen in Bangladesh, where 52% of E. coli isolates from extraintestinal tissues had blaCTX-M prevalence. Compared to the current study results, the study observed a decreased prevalence of blaTEM (20%) and blaOXA (17%) [21]. In addition, Results of genomic analysis showed blaTEM (100%) and blaCTX-M (16%) in the E. coli isolated from river water in Delhi, India [22]. According to findings of current and previous studies, there is a high prevalence of ESBLS in community and hospital acquired bacteria [23,24]
There is documented evidence of a rise in the predominant of antibiotic resistance in E. coli infections. Consequently, it is crucial to educate the local community about the appropriate utilization of antibiotics in order to counteract the rise in resistance [25-27]. Comprehending the genetic background of the virus is crucial to obtain adequate insights into mutations and resistance [28,29].
Conclusion
During therapy management, the gynecologists/urogynecologists physicians are concerned about the substantial spread of resistance mechanisms among the bacteria. Hospitalized and non-hospitalized people are experiencing a significant increase in multidrug resistant E. coli, which can lead to potentially fatal infections. The recent investigation showed that the E. coli isolates had many chromosomal and plasmid genes, which conferred resistance against third generation cephalosporins.
Funding
This study did not receive any funding.
Conflict of Interest
The authors declared there is no conflict of interest.
Authors’ Contribution
(A) Study Design · (B) Data Collection · (C) Statistical Analysis · (D) Data Interpretation · (E) Manuscript Preparation · (F) Literature Search · (G) No Fund Collection
References
- Mancuso G, Midiri A, Gerace E, et al. Urinary tract infections: The current scenario and future prospects. Pathogens. 2023; 12(4):623.
- Zhou Y, Zhou Z, Zheng L, et al. Urinary tract infections caused by uropathogenic E. coli: Mechanisms of infection and treatment options. Int J Mol Sci. 2023; 24(13):10537.
- Husna A, Rahman MM, Badruzzaman AT, et al. Extended-Spectrum Β-Lactamases (ESBL): challenges and opportunities. Biomedicines. 2023; 11(11):2937.
- Shaikh S, Fatima J, Shakil S, et al. Antibiotic resistance and extended spectrum beta-lactamases: Types, epidemiology and treatment. Saudi J Biol Sci 2015; 22(1):90-101.
- Noster J, Thelen P, Hamprecht A. Detection of multidrug-resistant enterobacterales—from ESBLs to carbapenemases. Antibiotics. 2021; 10(9):1140.
- Kim SW, Lee JS, Park SB, et al. The importance of porins and β-lactamase in outer membrane vesicles on the hydrolysis of β-lactam antibiotics. Int J Mol Sci. 2020; 21(8):2822.
- Fernández L, Hancock RE. Adaptive and mutational resistance: Role of porins and efflux pumps in drug resistance. Clin Microbiol Rev. 2012; 25(4):661-681.
- Kurazono H, Nakano M, Yamamoto S, et al. Distribution of the usp gene in uropathogenic E. coli isolated from companion animals and correlation with serotypes and size‐variations of the pathogenicity island. Microbiol Immunol. 2003; 47(10):797-802.
- Brolund A, Wisell KT, Edquist PJ, et al. Development of a real-time SYBRGreen PCR assay for rapid detection of acquired AmpC in Enterobacteriaceae. J Microbiol Methods. 2010; 82(3):229-233.
- Odoki M, Almustapha Aliero A, Tibyangye J, et al. Prevalence of bacterial urinary tract infections and associated factors among patients attending hospitals in Bushenyi district, Uganda Int J Microbiol. 2019; 2019(1):4246780.
- Silva A, Costa E, Freitas A, et al. Revisiting the frequency and antimicrobial resistance patterns of bacteria implicated in community urinary tract infections. Antibiotics. 2022; 11(6):768.
- Karlowsky JA, Kelly LJ, Thornsberry C, et al. Trends in antimicrobial resistance among urinary tract infection isolates of E. coli from female outpatients in the United States. Antimicrob Agents Chemother. 2002; 46(8):2540-2545.
- Asokan GV, Vanitha A. Disaster response under one health in the aftermath of Nepal earthquake, 2015. J Epidemiol Glob Health. 2017; 7(1):91-96.
- Susić E. Mechanisms of resistance in enterobacteriaceae towards beta-lactamase antibiotics. Acta Med Croatica. 2004; 58(4):307-312.
- Dirar Mh, Bilal Ne, Ibrahim Me, et al. Prevalence of Extended-Spectrum Β-Lactamase (ESBL) and molecular detection of bla TEM, bla SHV and bla CTX-M genotypes among Enterobacteriaceae isolates from patients in Khartoum, Sudan. Pan Afr Med J. 2020; 37(1).
- Zaniani FR, Meshkat Z, Nasab MN, et al. The prevalence of TEM and SHV genes among extended-spectrum beta-lactamases producing E. coli and K. pneumoniae. Iran J Basic Med Sci. 2012; 15(1):654.
- Kiratisin P, Apisarnthanarak A, Laesripa C, et al. Molecular characterization and epidemiology of extended-spectrum-β-lactamase-producing E. coli and Klebsiella pneumoniae isolates causing health care-associated infection in Thailand, where the CTX-M family is endemic. Antimicrob Agents Chemother. 2008; 52(8):2818-2824.
- Jain P, Bepari AK, Sen PK, et al. High prevalence of multiple antibiotic resistance in clinical E. coli isolates from Bangladesh and prediction of molecular resistance determinants using WGS of an XDR isolate. Sci Rep. 2021; 11(1):22859.
- Kandi V, Shahapur PR, Suvvari TK, et al. Molecular characterization of E. coli causing urinary tract infections through next-generation sequencing: A comprehensive analysis of serotypes, sequence types, and antimicrobial and virulence genes. Cureus. 2024; 16(3).
- Bush K, Bradford PA. β-lactams and β-lactamase inhibitors: An overview. Cold Spring Harb Perspect Med. 2016; 6(8):a025247.
- Mazumder R, Abdullah A, Ahmed D, et al. High prevalence of Bla CTX-M-15 gene among extended-spectrum β-lactamase-producing E. coli isolates causing extraintestinal infections in Bangladesh. Antibiotics. 2020; 9(11):796.
- Singh NS, Singhal N, Virdi JS. Genetic environment of bla TEM-1, bla CTX-M-15, bla CMY-42 and characterization of integrons of E. coli isolated from an Indian urban aquatic environment. Front Microbiol. 2018; 9:382.
- Ahjel SW, Hassan SM, Hussein SF, et al. Antineoplastic effect of new synthesized compounds of 2-thiouracil sulfonamide derivatives against ovarian and breast carcinoma cells" in vitro study". Sys Rev Pharm. 2020; 11(4).
- Aldulaimi AK, Idan AH, Majhool AA, et al. Synthesis of new antibiotic agent based on mannich reaction. Int J Drug Deliv Tec. 2022; 12(3):1428-1432.
- Wayne PA. Clinical and laboratory standards institute. Performance standards for antimicrobial susceptibility testing.
- http://www.latamjpharm.org/resumenes/0/6/LAJOP_0_6_1_22.pdf
- Ejaz H, Zafar A, Amin H. Multidrug resistant AmpC β-lactamase producing E. coli isolated from a paediatric hospital. Pak J Med Sci. 2014; 30(1):181.
- Pitout JD, Hossain A, Hanson ND. Phenotypic and molecular detection of CTX-M-β-lactamases produced by E. coli and Klebsiella spp. J Clin Microbiol. 2004; 42(12):5715-5721.
- Brinas L, Zarazaga M, Sáenz Y, et al. β-lactamases in ampicillin-resistant E. coli isolates from foods, humans, and healthy animals. Antimicrob Agents Chemother. 2002; 46(10):3156-3163.
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Author Info
Khamael Arif Mahdi Al-challabi1, Wurood Kadhim Abed2 and Ali Razzaq Hussein3*2Department of Medical Science, Jabir Ibn Hayyan University for Medical and Pharmaceutical Sciences, Al-Najaf, Iraq
3Directorate of Education in Al-Najaf Ministry of Education, Al-Najaf, Iraq
Copyright:This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.